Intro
In this (belated) post I summarise and extend my previous essay about human-chimpanzee genome alignments and how they’ve recently been interpreted by some opponents of evolutionary theory, particularly as reported at “Evolution News” from the Discovery Institute, but now picked up on by many others. There is a huge amount of data in the scientific paper that was the basis of all of this so I’ve just highlighted some of it, which took me long enough! The main novelty in this second post from me, in relation to the broader discussion happening elsewhere, is investigating the likely contribution of alignment failure to the magic 14.9% number.
Dr. Casey Luskin has now written a series of responses to the various critics of the Evolution News articles, including me. You can read them to see how they compare to what I’ve written. Responding to the full set of 13 articles at Evolution News, ‘Gutsick Gibbon’ recently released a long response video, so do go to that if you want more. She makes a lot of really good points and I agree with most of what she says in the parts I listened to, though I think she was overly critical in a couple of places, positing blatant dishonesty when I think what’s going on at some points is a bit more subtle. I agree however that failing to provide some of the key context for understanding the numbers such as differences within the human species and between other species is definitely problematic. There is more interesting discussion at Peaceful Science, too.
The TL;DR version: many non-aligning regions are demonstrably repetitive regions which most probably cannot be aligned with the standard algorithms but are not composed of new sequence - they have not been shown to be differences, let alone functional differences. The claim that human and chimpanzees differ by ~14.9% is in my assessment not supported by the evidence. As a post at Evolution News helpfully says, “at least get the facts straight”. More importantly though, the purported implications of the various statistics available have been mis-stated - the degree of difference observed is typical across mammalian families - a taxonomic level where prominent proponents of Intelligent Design and various strains of creationism accept that common ancestry holds true and evolutionary mechanisms explain observed differences.
Note, I’m not here to defend a 1% overall genome difference between humans and chimps. Already in 2005 the difference was claimed to be 4% (not 1%) and how accurate this still is today will depend on exactly what we’re counting, as explained in my previous post and more below.
Recap
A scientific paper published in Nature (Yoo et al.) has recently reported complete genome sequences for six non-human ape species, and comparisons among them and to previously assembled complete human genomes. In my previous substack post on this topic, I made four main points regarding the interpretation presented by evolution skeptics at “Evolution News”. I still believe these points were correct! Here I restate them more clearly and concisely than before:
(1) Some genome regions can’t align well for technical reasons due e.g. to highly repetitive sequences. This means that aside from the actual biological meaningfulness and supposed implications of any numbers given, discussed below, the empirical claim of at least 14.9% sequence difference presented across multiple posts at “Evolution News” (and picked up by various anti-evolution sources) is not justified by the evidence presented.
(2) Copy number variants or repeats aren’t differences in the same way as genuinely new sequences are. Sometimes copy number variants or repeat regions are functionally important but they should be distinguished from other variants as they aren’t new sequences. This is a point which many opponents of evolution are keen to press in other contexts.
(3) There are large differences within the human species and within the chimpanzee species (as for other mammalian species) which aren’t considered at all. Within-group differences are highly relevant to claims about the magnitude of between-group differences. Contra “Evolution News”, this is not a case of “Try to change the subject and talk about something else”.
(4) Nothing was presented to indicate that the human-chimp (and other ape) differences are different to what is seen across other mammalian families. This is problematic for the narrative that the findings have implications for common ancestry, human origins, or human exceptionalism, let alone all three as was claimed in the posts at Evolution News. This point hasn’t been addressed in the responses and isn’t one I’ve seen any coherent response to from evolution skeptics. The uncomfortable fact is that the human-chimpanzee genomic divergence falls within the range of divergences that most informed evolution skeptics are happy to accept as arising from common ancestry of mammalian Families (including young Earth creationists who have to cram this divergence into a smaller time period).
Developing these points further:
(1) I develop this point in most depth as no-one else has investigated this, that I’ve seen. Evolution-skeptic Dr. Casey Luskin has said e.g. “There are huge portions of the human and chimp genomes that are so different that they simply cannot be aligned and compared” and “12.5 percent to 14 percent of the genomes are so different that they can’t even be aligned to make a comparison”, and that there is “12.5 percent to 14 percent that are completely different”. “Completely different” is clearly wrong - he writes elsewhere that many of the differences are in repetitive elements i.e. they are duplicated regions rather than completely different stretches of sequence. But “so different that they can’t even be aligned” is also likely an incorrect framing - sequences needn’t be different in order to not be alignable.
Non-aligning regions in a pairwise genome comparison are often repetitive sequences - which may be present in both genomes but can’t align well because of the way aligners work. As one reference for a similar tool to one of the alignment methods used in Yoo et al. puts it, “Minigraph-Cactus (in common with all multi-sequence alignment tools that we know of) cannot presently satisfactorily align highly repetitive sequences, such as satellite arrays, centromeres and telomeres, because they lack sufficiently unique subsequences for minigraph to use as alignment seeds.” As such, gap differences can’t be assumed to be biological differences. Gap differences due to alignment failure are seen even when comparing human genomes against each other. Dr. Casey Luskin has attempted to respond to this but I believe missed the point.
After my previous substack post I’ve been asked to note that in his essays Casey has repeatedly said that repeats are a big part of the difference between human and chimpanzee sequence. This is true - though it undermines his other claims about the 14.9% gap divergence representing completely different sequences, and the fact that copy number variants are not differences in information content in the usual sense was in my view downplayed. In any case, the main two issues with repetitive sequence regions that I raised were not that they’re “junk” (though that may well be true for much of it, a topic for another day) but that they’re differences as implied in some of the wording at Evolution News and that a sizeable proportion of it won’t align simply because of how aligners work. Because I irrationally care too much about getting this stuff right, I checked the human vs chimp genome alignments provided alongside the Yoo et al. paper (my code is provided for reproducibility at the end of this post - it should take approx 2hours to run everything).
What percentage of the human genome lacks an aligned sequence from chimpanzee (from the wfmash alignments)? I calculate it as 10% (290771270 gaps / 2900555906 total autosome length). This is a bit lower than some of the other percentages reported in the Yoo et al. paper (based on other genomes and/or medians of 1Mb chunks), but that just makes this analysis more conservative and the difference shouldn’t matter for the broad brush strokes I paint below.
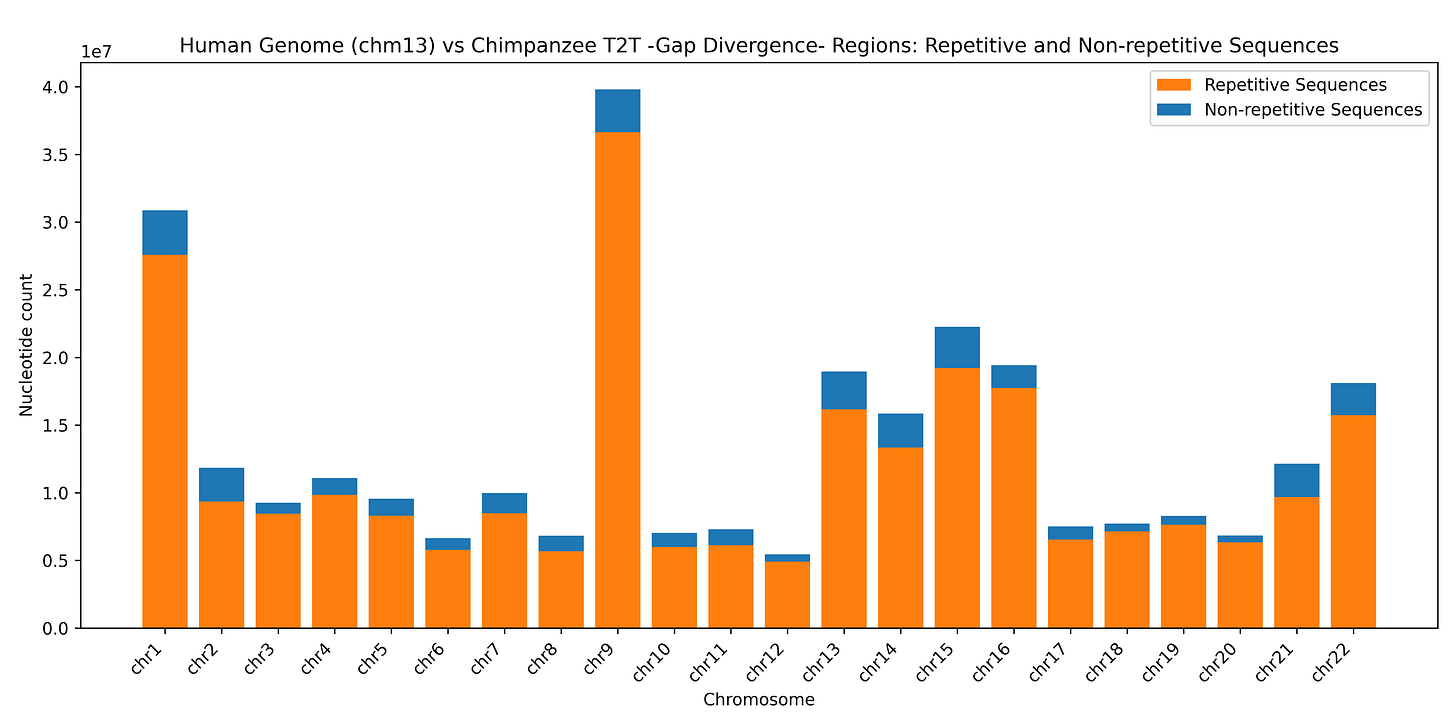
How much of the “gap divergence” is associated with repetitive regions? By far the most of it. Most sequences with gaps in the chimp sequence relative to the full T2T human sequence (i.e. human sequences that evolution skeptics are interpreting as unique to humans - “completely different”) are highly repetitive regions in the human reference (Figure 1), “soft masked” by the program RepeatMasker as lowercase letters (as far as I can understand the files available) - regions where it is well known that aligners don’t work well.
Figure 1: most “gap divergence” regions of the human genome (regions without alignment to the chimpanzee genome) are repetitive sequences, indicated with lowercase letters in the MAF format alignment files created using wfmash.
Here are 20 random examples of “gap divergence” regions which are lowercase sequences (i.e. soft-masked to indicate repeat regions), trimmed to 100 nucleotides, to confirm these are indeed typically repeat regions:
1.ggagatcgagaccatcctggctaacaaggtgaaaccccgtctctactaaaaatacaaaaaattagccgggcgcggtggcg
2.ataaatttcatatattatatataaatataagtatatattatatataaatttcatatagtatatataaatataattatata
3.tttttttttttttgaaacggagtcttgctctgtcgcccaggctggagtgcagtggcgcgatctcagctcactgtaagctc
4.ttttttttttttttttttgagacggagtctcgctctgtcgcccaggccggactgcggactgcagtggcgcaatctaggct
5.aacccgggaggcggagcttgcagtgagtcgagatcgcgccactgcactccagcctgggcgacagagcaaaactccgcctc
6.ctgcactcaagcctggggtgacagagtgagatcctgtccaaaaaaaaaataaaacaacaacaacaaaacaaacaatagaa
7.tatatatatatatatatatatatatagagagagagagagagagagagagagagagagagagagagagagagagagagaga
8.ttttagtagagacggggtttcaccgtgttagccaggctggtctcgatctcctgacctcgtgatccgcccgcctcggcctc
9.tttgggttggttccaagtctttgctattatggataatgccacaataaacatacgtgtgcatgtgtctttatagcagcatg
10.agggcgcctgtagtcccagctacacgggaggctgaggcaggagaatggggtgaacccgggaggcggagcttgcagtgagt
11.tccccaatgttttcttgtagtaattttatagtacaagatcttagctataagtctttaatccattttgactttagttttgt
12.acctaatgctagatgacacattagtgggtgcagcacaccagcatggcacatgtatatatatgtaactaacctgcacaatg
13.acctgcagtgggtagctgctgtcttcaggcaggtcatctcatctcatctcatctctgcaaccctcagcaaagaagagata
14.gggtggatcatgaggtcagtagatcgagaccatcctggctaacaaggtgaaaccccgtctctactaaaaatacaaaaaat
15.tgtgggttgtctgtttactctgctgactgttccttttgctgtgcaaacactttctagtttaattaagtcccagctattta
16.atttcccagtctggagtatgttcttccttacaggcttctatggcattctgtttcctctatgatagctctaatggttttta
17.tatttgactgggccacagggtgtccagacatttggctgaatattaatctgagtgtgtctgtgagggtgtttctgggtgag
18.tacacatatatacacatatacacatatatatacatatatacacatatatatacacatatatatatacacatatatatgtg
19.accaccacaccatcaccatcatcaccaccacacccatcaccaccatcaccaccaccacacccatcaccaccatcatcacc
20.atatatattcctatacatatattcctatatatattcctatatatattcctatacatatacattcctatatatatattcctI ran this through ChatGPT for a summary table of the repeat elements seen in these sequences, but the details are irrelevant and somewhat boring - you can see many of the repeats by eye, and if not convinced you can ask chatGPT yourself! Some of the repeats are probably longer and so missed in these short snippets, but I haven’t investigated.
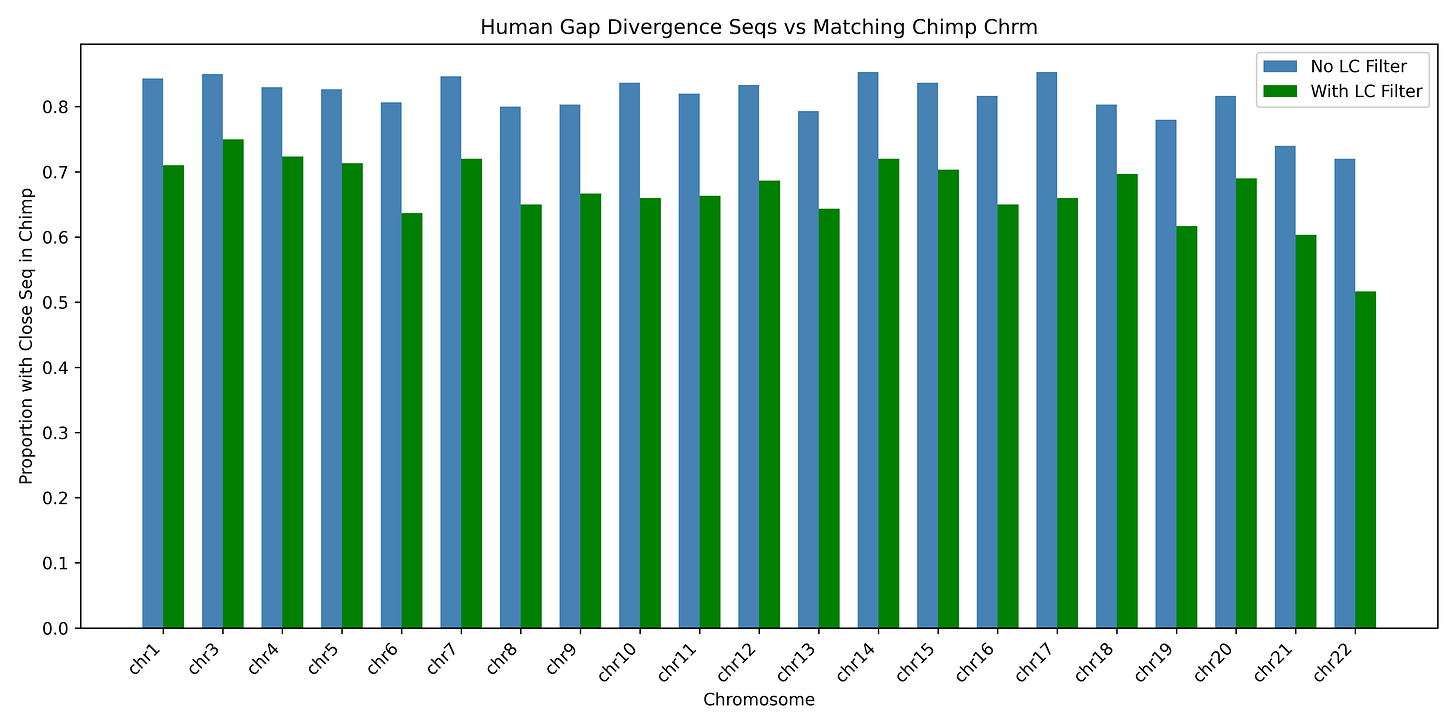
Okay, so the “gap divergence” regions are mostly highly repetitive sequences, but maybe they’re still different to what’s found in the chimpanzee genome. So, are these non-aligning repeat elements, though found in many copies in the human genome, unique to humans? No - most of the “gap divergence” sequences from the human genome have similar or identical sequences found in the chimpanzee genome, in the chromosome which aligns best to the human chromosome they’re found in. To test this, from each human (CHM13) chromosome’s list of “gap divergence” regions I chose 300 random sequences of 100 nucleotides long, and queried them (BLASTn) against the corresponding chimpanzee chromosome, conservatively requiring an alignment length of at least 80 and percentage identity of 80 to count as a match. The large majority have a good match. (I excluded chromosome 2 just as it’s more complicated to deal with due to the fusion).
Figure 2: most “gap divergence” regions of the human genome (regions without alignment to the chimpanzee genome) have at least one close match in the equivalent chimpanzee chromosome - regardless of whether a low-complexity filter is used in the BLAST search (chromosome 2 excluded just to avoid dealing with the fusion).
That seems like enough to kill the claim that human and chimpanzee genomes are actually ~15% different, if different means that the “gap divergence” sequences are present in the human but not the chimpanzee genome. To remind you, Casey Luskin said “At least 12.5 percent … represent a “gap difference” between the two genomes. That means there’s a “gap” in one genome compared to the other, often where they are so different, they cannot even be aligned.” This implies that failure to align, due to major sequence difference, is the norm - but most human “gap divergence” regions in fact usually have a close match (or many) in the appropriate chimp chromosome.
However, perhaps all of these repeat regions - while the majority are clearly not sequence motifs unique to humans - are copy number variants where the number of copies is higher in (this particular) human than (this particular) chimpanzee genome. i.e. repeats unique to (this particular) human as compared to the chimpanzee. It turns out this isn’t the case (Figure 3) - the majority of these sequences have close matches in the parts of the chimpanzee genome that don’t align to the human genome (the gap divergence regions when doing the pairwise alignment in reverse). This strongly supports my suggestion that a lot of the supposed differences in the “gap divergence” regions are due to technical failures of alignment in repetitive regions rather than actual sequence differences.
Figure 3: most “gap divergence” regions of the human genome (regions without alignment to the chimpanzee genome) have at least one close match in the “gap divergence” regions of the alignment of the equivalent chimpanzee chromosome.
Even aligning two haplotypes of the same genome (HG002) against each other shows a mean of 3.48 gap divergence and 0.16% SNP divergence across the windows used (autosomes only), for a total difference of 3.64% (sum the values from this table). Or, by a slightly different measure, gaps comprise in total 4.29% of the alignment (I haven’t tried to work out why these values differ). This data comes from a sequenced mother, father, son trio, with Ashkenazi Jewish ethnic background - the parents should be quite genetically similar to each other as compared to randomly chosen humans, given the history of that community. Presumably, as with the human-chimp comparison - a large proportion of the gaps do not represent genuinely missing/novel sequences. I leave further investigation of these things to the reader!
After this overly long technical exploration I conclude that the 14.9% difference claim is unsupported due to misunderstanding the technical difficulties with aligning repetitive regions, quite apart from the more important issues of interpretation and presentation of the alignment statistics, which I discuss briefly below.
(2) Copy number variants are a different kind of difference to genuinely new sequences.
Changes in sequence copy number or the number of various types of repetitive elements are very common in the human genome, occurring between parents and children (there are lots of studies investigating this topic). This is a very good reason to be cautious about using such variation as a relevant measure of distance between human and chimpanzee genomes. As Joel Duff has noted (I think in this video but can’t find it now …), an example of this phenomenon - variable number tandem repeats - is robust enough to be used in forensics to identify individuals. This kind of difference doesn’t fit the wording at ‘Evolution News’ or in anti-evolution sites picking up this story, and the massive differences in such regions within the human population (not to mention within other groups that anti-evolutionists accept share common ancestry) undermine their purported relevance as somehow undermining the genetic case for common ancestry.
(3) Within species differences are highly relevant to claims of between species differences
This general issue is seen across fields in “within group” vs “between group” comparisons of variance. One way that the importance of within-species differences to between-species differences is clearly demonstrated in the data from Yoo et al is seen by comparing Supplementary Table III.17 to Supplementary Table III.19. Evolution News cites the human-chimp similarity from Table 19, i.e. 84.95% similarity (15.05% difference). When the chimpanzee pangenome is used (i.e. known within-species differences - a subset of actual differences - are included for chimpanzee), the percentage of the human autosomes covered by exactly identical chimpanzee sequence with a 1:1 alignment increases to 87.78 (12.22% difference). Many of these putative species differences will likewise be found within the human pangenome, and others are probably due to technical limitations of alignment in highly repetitive regions, as we’ve seen.
These considerations don’t even consider the much larger group that the author of the Evolution News posts counts as “human” - which includes Neanderthal and Denisovans. Others in the Intelligent Design movement and many young Earth creationists (who have jumped on board the 14.9% difference train) would include Homo erectus, the genomes of which are of course expected to differ even more from modern humans than Neanderthal and Denisovan genomes do. This simple appealing claim that humans and chimpanzees are ~15% different genetically is starting to look like a bit of a headache for those opposing evolution ...
(4) Differences calculated with telomere to telomere assemblies will increase regardless of what is compared
Fundamentally, comparing the percentage of sequence aligning to the SNP-based differences often reported in the popular literature is mistaken, as multiple critics have pointed out - it is comparing apples with oranges. Further, if you use the newly available full (telomere-to-telomere) genome assemblies and count all non-aligning regions as differences, the “differences” seen within any mammalian family will increase as compared to using the previous generation of assemblies without some of the added highly repetitive regions. Genomic differences within other mammalian groups which most anti-evolutionists are happy to accept share common ancestry are similar to those seen within the taxonomic group of apes (including humans). This basic but somewhat awkward context completely undercuts the remarkable fuss that the Discovery Institute have decided to try to make about this Nature paper on new ape genome assemblies.
Summary
This post critiques claims by some evolution skeptics, particularly at the Discovery Institute's Evolution News, that humans and chimpanzees differ by ~14.9% at the genome level. These non-aligning regions are often actually shared between species but are difficult to compare due to technical limitations of alignment algorithms, not biological novelty. Copy number variations and alignment failures are being presented by evolution critics as real novel sequences. Furthermore, similar degrees of variation exist within species and across other mammalian families, undermining claims about human exceptionalism or the breakdown of evolutionary theory. My critique particularly emphasizes the importance of (1) distinguishing between technical artifacts and the various kinds of potentially meaningful biological differences in genome comparisons and (2) providing the important biological context of differences within the species and within other mammalian groups.
Final Comment - Errors
Genome alignment is a very technical field, so it’s easy to get details wrong. Adding in considerations of pangenomes and within-species differences gets even more complex. My expertise is primarily in microbial genomes so I’m not familiar with some of the features of large eukaryote genomes. No one likes being wrong, but I want to be open to being corrected so do reach out to me if you think you see a mistake in fact or in interpretation. If you’re reading this and involved in producing or promoting some of the content for any of the various viewpoints available, you could play a part in shifting the discussion to one where accurate science trumps the rhetoric and political posturing.
I’ve made some errors along the way in my explorations of this in my spare time over the last few weeks, although nothing significant I think in the previous blog post. In particular, the chimpanzee genome is quite a bit longer than the human genome and for some time I thought this was a factor in the percentage differences - it’s likely not, as every relevant statistic in the Yoo et al. paper is effectively a proportion of the human genome covered by a similar chimpanzee sequence. It is also easy to not properly understand the differences between the various pairwise comparisons listed in the Yoo et al. paper. This isn’t due to some conspiracy of genome scientists or journals, but just because the field is highly technical and the interest of specialists differs from that of the general public, particularly members of the public who are skeptical of evolutionary theory and don’t operate within the framework that the vast majority of genomics researchers work within.
I note that similar claims to those currently circulating have been made for many years by young Earth creationists (as explored e.g. in many videos from biological anthropology PhD student Erika a.k.a. “Gutsick Gibbon” and biology professor Dr. Joel Duff). Key lessons from this previous work have unfortunately been missed by skeptics of evolution who should be paying attention to such things.
Thanks for reading this overly long post and I hope it has helped you understand comparative genomics a bit better.
Code
If you want to reproduce what I’ve done (as unlikely as that is!) this overly long code below will be a good start. You can run it in chunks, and not all of the initial downloads will be necessary. Feel free to reach out to me if I’ve missed something out or made a significant mistake.
#!/bin/bash
###### PROGRAMS required, other than basic gnu/linux tools:
# aws
# gawk (should be faster than awk but can use awk if preferred)
# python3 with various packages installed
# BLAST+ from NCBI
###### DOWNLOAD FILES
wget https://garrisonlab.s3.amazonaws.com/t2t-primates/wfmash-v0.13.0/alignments_fixedSiamang/mPanTro3%231.p70.aln.paf.gz ;
NAMES=( "chm13#1" \
"hg002#M" "hg002#P" \
"mPanTro3#1" "mPanTro3#2" \
"mPanPan1#M" "mPanPan1#P" ) ;
mkdir mafs
cd mafs ;
# ALIGNMENTS AGAINST CHM13 T2T HUMAN GENOME
mkdir chm13#1 ;
cd chm13#1 ;
aws s3 cp s3://garrisonlab/t2t-primates/wfmash-v0.13.0/mafs_fixedSiamang/ . --no-sign-request --recursive --exclude "*" --include "chm13#1*filtered10Mb.maf.gz" ;
rm *chrM* ;
cd .. ;
# ALIGNMENTS AGAINST HG002 T2T HUMAN GENOME - MATERNAL HAPLOTYPE
mkdir hg002#M
cd hg002#M
aws s3 cp s3://garrisonlab/t2t-primates/wfmash-v0.13.0/mafs_fixedSiamang/ . --no-sign-request --recursive --exclude "*" --include "hg002#M*filtered10Mb.maf.gz"
rm *chrM* ;
cd .. ;
# ALIGNMENTS AGAINST 'CLASSIC' GRCH38 STANDARD REFERENCE HUMAN GENOME
mkdir grch38#1
cd grch38#1
aws s3 cp s3://garrisonlab/t2t-primates/wfmash-v0.13.0/mafs_fixedSiamang/ . --no-sign-request --recursive --exclude "*" --include "grch38#1*filtered10Mb.maf.gz"
rm *chrM* ;
cd .. ;
# ALIGNMENTS AGAINST CHIMPANZEE (HAPLOTYPE 1)
mkdir mPanTro3#1
cd mPanTro3#1
aws s3 cp s3://garrisonlab/t2t-primates/wfmash-v0.13.0/mafs_fixedSiamang/ . --no-sign-request --recursive --exclude "*" --include "mPanTro3#1*filtered10Mb.maf.gz"
rm *chrM* ;
cd .. ;
# note - folder with key alignment (maf) files from wfmash aligner
# https://garrisonlab.s3.amazonaws.com/index.html?prefix=t2t-primates/wfmash-v0.13.0/mafs_fixedSiamang/
##### IN ALIGNMENT: COUNT REPEAT SEQUENCES (HUMAN-REF - CHM13) AND GAPS (CHIMP)
# time: ~3m50s
cd chm13#1 ;
time for maf_file in *maf.gz ; do
chr=$(echo $maf_file | awk -F "[#|.]" '{print $3}') ;
echo $chr ;
if [[ "$chr" != "chrX" && "$chr" != "chrY" && "$chr" != "chr2" ]]; then
mkdir $chr ; cd $chr ;
gzcat ../"$maf_file" | gawk '$2 ~ "chm13#1" { print $7 > "hum_seq.txt" } $2 ~ "PanTro3#1" { print $7 > "chimp_seq.txt" }' ;
for seq_file in hum_seq.txt chimp_seq.txt ; do
python -c "from collections import Counter; import sys; counts = Counter(open(sys.argv[1]).read()); counts.pop('\n', None); [print(f'{v} {c}') for c, v in counts.most_common()]" "$seq_file" > "${seq_file%.*}"_counts.txt ;
done ;
cd .. ;
fi ;
if [[ "$chr" == "chr2" ]]; then
mkdir $chr ; cd $chr ;
gzcat ../"$maf_file" | gawk '$2 ~ "chm13#1" { print $7 > "hum_seq.txt" } $2 ~ "PanTro3#1" { print $7 > "chimp_seqs.txt" }' ;
python -c "from collections import Counter; import sys; counts = Counter(open(sys.argv[1]).read()); counts.pop('\n', None); [print(f'{v} {c}') for c, v in counts.most_common()]" hum_seq.txt > hum_seq_counts.txt ;
# for chrm 2 only count gaps if gaps across both chimp seqs; otherwise count the nucleotide present
python3 -c "import sys; from collections import Counter; seq1, seq2 = open(sys.argv[1]).read().splitlines(); counts = Counter(); [counts.update(['-']) if a=='-' and b=='-' else counts.update([b]) if a=='-' else counts.update([a]) if b=='-' else (_ for _ in ()).throw(ValueError(f'No gap at position {i+1}: got {a} and {b}')) for i, (a,b) in enumerate(zip(seq1, seq2))]; [print(f'{v} {k}') for k,v in counts.most_common()]" chimp_seqs.txt > chimp_seq_counts.txt
cd .. ;
fi ;
done ;
##### COUNT PROPORTION OF "GAP DIVERGENCE" SEQS (PRESENT IN HUMAN, NOT MATCHED TO CHIMP) WHICH ARE IN REPETITIVE REGIONS
# time: ~40s
time for maf_file in *maf.gz ;
do
chr=$(echo $maf_file | awk -F "[#|.]" '{print $3}') ;
echo $chr ;
# chr2 is special case where there are two seqs (because of fused chromosome in relation to humans); both must be gaps to be counted as a gap
if [[ "$chr" == "chr2" ]]; then
cd $chr ;
python3 -c "import re, sys; seq1, seq2 = open('chimp_seqs.txt').read().splitlines(); \
chr = sys.argv[1]; \
both = ''.join('-' if a == b == '-' else 'X' for a,b in zip(seq1, seq2)); \
[print(f'{chr}\t{m.start()}\t{m.end()}') for m in re.finditer(r'-+', both)]" "$chr" > chimp_seq_gap_coords.txt ;
cd .. ;
fi ;
if [[ "$chr" != "chrX" && "$chr" != "chrY" && "$chr" != "chr2" ]]; then
cd $chr ;
python3 -c "import re, sys; seq = open('chimp_seq.txt').read(); \
chr = sys.argv[1]; \
[print(f'{chr}\t{m.start()}\t{m.end()}') for m in re.finditer(r\"-+\", seq)]" "$chr" > chimp_seq_gap_coords.txt ;
cd .. ;
fi ;
if [[ "$chr" != "chrX" && "$chr" != "chrY" ]]; then
cd $chr ;
python3 -c "import sys; hum_seq = open(sys.argv[1]).read(); lower_count = 0; upper_count = 0; \
f_coords = open(sys.argv[2]); f_out = open(sys.argv[3], 'w'); \
[ (lambda parts: \
[ f_out.write(f'{hum_seq[int(parts[1]):int(parts[2])+1]}\\n'), \
globals().__setitem__('lower_count', lower_count + sum(1 for c in hum_seq[int(parts[1]):int(parts[2])+1] if c.islower())), \
globals().__setitem__('upper_count', upper_count + sum(1 for c in hum_seq[int(parts[1]):int(parts[2])+1] if c.isupper())) ])(line.strip().split()) for line in f_coords ]; \
f_coords.close(); f_out.close(); \
f_counts = open(sys.argv[4], 'w'); \
f_counts.write(f'Total lowercase letters: {lower_count}\\n'); \
f_counts.write(f'Total uppercase letters: {upper_count}\\n'); \
f_counts.close()" hum_seq.txt chimp_seq_gap_coords.txt hum_gap_regions.txt letter_case_counts.txt ;
cd .. ;
fi ;
done ;
# PLOT FIGURE 1
python3 -c "
import matplotlib.pyplot as plt
import glob, re
data = []
for d in glob.glob('chr*'):
if d in ('chrX', 'chrY'): continue
with open(f'{d}/letter_case_counts.txt') as f:
txt = f.read()
lower = int(re.search(r'Total lowercase letters: (\d+)', txt).group(1))
upper = int(re.search(r'Total uppercase letters: (\d+)', txt).group(1))
data.append((d, lower, upper))
# Sort numerically by chromosome number
data.sort(key=lambda x: int(re.sub(r'chr', '', x[0])))
chroms = [x[0] for x in data]
lower = [x[1] for x in data]
upper = [x[2] for x in data]
plt.figure(figsize=(12, 6))
# Keep stacking order: lower then upper
# But change colors: lower = bright orange, upper = dark blue
bars1 = plt.bar(chroms, lower, label='Repetitive Sequences', color='#ff7f0e')
bars2 = plt.bar(chroms, upper, bottom=lower, label='Non-repetitive Sequences', color='#1f77b4')
plt.ylabel('Nucleotide count')
plt.xlabel('Chromosome')
plt.title('Human Genome (chm13) vs Chimpanzee T2T -Gap Divergence- Regions: Repetitive and Non-repetitive Sequences')
plt.xticks(rotation=45, ha='right')
plt.legend()
plt.tight_layout()
plt.savefig('Fig1.png', dpi=600)
plt.show()
"
# GET 20 RANDOM SEQUENCES FROM THE GAP DIVERGENCE REGIONS, ONLY CONSIDERING SOFT-MASKED REGIONS:
echo "20 GAP DIVERGENCE SEQS, EXAMPLES" ;
for chr_folder in chr*; do
cat "$chr_folder/hum_gap_regions.txt" ; done |
awk 'length($0) >= 80' |
awk 'BEGIN { srand() } { start = int(1 + rand() * (length($0) - 80 + 1)); s = substr($0, start, 80); if (s ~ /^[a-z]+$/) print s}' | shuf -n 20 ;
# SEARCH 300 RANDOM SEQUENCES FROM THE GAP DIVERGENCE REGIONS AGAINST THE CORRESPONDING CHIMPANZEE CHROMOSOMES
# prep chimp chromosome data (already downloaded)
# time: approx 15mins
time for maf_file in *maf.gz ;
do
hum_chr=$(echo $maf_file | awk -F "[#|.]" '{print $3}') ;
echo $hum_chr ;
hsa=$(echo $hum_chr | sed -e "s|chr|hsa|g") ;
echo $hsa ;
if [[ "$hum_chr" != "chrX" && "$hum_chr" != "chrY" && "$hum_chr" != "chr2" ]]; then
cd $hum_chr ;
gzcat ../../mPanTro3#1/*"$hsa".filtered10Mb.maf.gz | awk '{if ($2~"mPanTro3#1") print ">""'$hsa'" "\n" $7}' > chimp_full_seq_H1.fna ;
cd .. ;
fi ;
done ;
# BLASTn search gap divergence example seqs (from human) against corresponding full chimp chromosome
time for maf_file in *maf.gz ;
do
hum_chr=$(echo $maf_file | awk -F "[#|.]" '{print $3}') ;
if [[ "$hum_chr" != "chrX" && "$hum_chr" != "chrY" && "$hum_chr" != "chr2" ]]; then
echo $hum_chr ;
cd $hum_chr ;
cat hum_gap_regions.txt | awk 'length($0) >= 100' |
awk 'BEGIN { srand() } { start = int(1 + rand() * (length($0) - 100 + 1)); s = substr($0, start, 100); if (s ~ /^[a-z]+$/) print s}' | shuf -n 300 |
awk '{print ">"NR "\n" $1 }' > ran_gapdiv_qseqs.fna ;
makeblastdb -in chimp_full_seq_H1.fna -dbtype nucl -out chimp_full_seq_H1 ;
blastn -query ran_gapdiv_qseqs.fna -db chimp_full_seq_H1 -out blast_gapdiv_results1.txt -outfmt "6 qseqid sseqid pident length evalue bitscore qstart qend sstart send" -dust no ;
blastn -query ran_gapdiv_qseqs.fna -db chimp_full_seq_H1 -out blast_gapdiv_results2.txt -outfmt "6 qseqid sseqid pident length evalue bitscore qstart qend sstart send" -dust yes ;
cat blast_gapdiv_results1.txt | awk '{if ($4>80 && $3>80 && $5<1e-20) print $1}' | sort | uniq -c | awk '{print $1 "\t" $2}'> blast_gapdiv_results_counts1.txt ;
cat blast_gapdiv_results2.txt | awk '{if ($4>80 && $3>80 && $5<1e-20) print $1}' | sort | uniq -c | awk '{print $1 "\t" $2}' > blast_gapdiv_results_counts2.txt ;
cd .. ;
fi ;
done ;
# - FIG 2
# PLOT BLASTn RESULTS (human gap divergence seqs vs chimp chrm)
python3 -c """
import os, glob, numpy as np, matplotlib.pyplot as plt
folders = sorted([f for f in glob.glob('chr*') if f != 'chr2'],
key=lambda s: int(''.join(filter(str.isdigit, s)) or 1e9))
chroms, counts1, counts2 = [], [], []
for f in folders:
try:
c1 = sum(1 for _ in open(f'{f}/blast_gapdiv_results_counts1.txt'))
c2 = sum(1 for _ in open(f'{f}/blast_gapdiv_results_counts2.txt'))
chroms.append(f)
counts1.append(c1/300)
counts2.append(c2/300)
except FileNotFoundError:
continue
x = np.arange(len(chroms))
w = 0.35
fig, ax = plt.subplots(figsize=(12,6))
ax.bar(x - w/2, counts1, w, label='No LC Filter', color='steelblue')
ax.bar(x + w/2, counts2, w, label='With LC Filter', color='green')
ax.set(xlabel='Chromosome', ylabel='Proportion with Close Seq in Chimp',
title='Human Gap Divergence Seqs vs Matching Chimp Chrm',
xticks=x, xticklabels=chroms)
plt.xticks(rotation=45, ha='right')
ax.legend()
plt.tight_layout()
plt.savefig('Fig2.png', dpi=600)
plt.show()
"""
cd ..
# CHECK HOW MANY HUM GAP DIVERGENCE SEQS HAVE MATCH IN CHIMP GAP DIVERGENCE SEQS (SAME CHROMOSOME)
# EXTRACT CHIMP GAP DIVERGENCE SEQS
cd mPanTro3#1 ;
# time: 1m20s
time for maf_file in *maf.gz ;
do
chimp_chr=$(echo $maf_file | awk -F "[#|.|_]" '{print $3}') ;
hsa=$(echo $maf_file | awk -F "[_|.]" '{print $3}') ;
if [[ "$chimp_chr" != "chrX" && "$hsa" != "hsa2a" && "$hsa" != "hsa2b" ]]; then
echo $chimp_chr $hsa ;
test ! -d $hsa && mkdir $hsa ; cd $hsa ;
gzcat ../"$maf_file" | gawk '$2 ~ "chm13#1" { print $7 > "hum_seq_subject.txt" } $2 ~ "mPanTro3#1" { print $7 > "chimp_seq_ref.txt" }' ;
python3 -c "import re, sys; seq = open('hum_seq_subject.txt').read(); \
chr_subject = sys.argv[1]; \
[print(f'{chr_subject}\t{m.start()}\t{m.end()}') for m in re.finditer(r\"-+\", seq)]" "$hsa" > humsubject_seq_gap_coords.txt ;
python3 -c "import sys; s=open(sys.argv[1]).read(); o=open(sys.argv[3],'w'); \
[o.write(s[int(a):int(b)] + '\n') for _,a,b in (line.split() for line in open(sys.argv[2]))]; o.close()" chimp_seq_ref.txt humsubject_seq_gap_coords.txt chimp_gap_regions.txt
cat chimp_gap_regions.txt | awk '{if (length($1)>=80) print ">seq"NR"\n"$1 }' > chimp_gap_regions_long.fna ;
cd .. ;
fi ;
done ;
cd .. ;
# BLAST HUMAN GAP DIVERGENCE SEQ EXAMPLES AGAINST CHIMP GAP DIVERGENCE SEQS
cd chm13#1/ ;
time for maf_file in *maf.gz ;
do
hum_chr=$(echo $maf_file | awk -F "[#|.]" '{print $3}') ;
hsa=$(echo $hum_chr | sed -e "s|chr|hsa|g") ;
if [[ "$hum_chr" != "chrX" && "$hum_chr" != "chrY" && "$hum_chr" != "chr2" ]]; then
echo $hum_chr ;
cd $hum_chr ;
makeblastdb -in ../../mPanTro3#1/"$hsa"/chimp_gap_regions_long.fna -dbtype nucl -out chimp_gapdiv_longseqs_H1 ;
blastn -query ran_gapdiv_qseqs.fna -db chimp_gapdiv_longseqs_H1 -out blast_gapdiv_results3.txt -outfmt "6 qseqid sseqid pident length evalue bitscore qstart qend sstart send" -dust no ;
blastn -query ran_gapdiv_qseqs.fna -db chimp_gapdiv_longseqs_H1 -out blast_gapdiv_results4.txt -outfmt "6 qseqid sseqid pident length evalue bitscore qstart qend sstart send" -dust yes ;
cat blast_gapdiv_results3.txt | awk '{if ($4>80 && $3>80 && $5<1e-20) print $1}' | sort | uniq -c | awk '{print $1 "\t" $2}'> blast_gapdiv_results_counts3.txt ;
cat blast_gapdiv_results4.txt | awk '{if ($4>80 && $3>80 && $5<1e-20) print $1}' | sort | uniq -c | awk '{print $1 "\t" $2}' > blast_gapdiv_results_counts4.txt ;
cd .. ;
fi ;
done ;
# - FIG 3
# PLOT BLASTn RESULTS (human gap divergence seq examples vs all non-short chimp gap divergence seqs)
python3 -c """
import os, glob, numpy as np, matplotlib.pyplot as plt
folders = sorted([f for f in glob.glob('chr*') if f != 'chr2'],
key=lambda s: int(''.join(filter(str.isdigit, s)) or 1e9))
chroms, counts1, counts2 = [], [], []
for f in folders:
try:
c1 = sum(1 for _ in open(f'{f}/blast_gapdiv_results_counts3.txt'))
c2 = sum(1 for _ in open(f'{f}/blast_gapdiv_results_counts4.txt'))
chroms.append(f)
counts1.append(c1/300)
counts2.append(c2/300)
except FileNotFoundError:
continue
x = np.arange(len(chroms))
w = 0.35
fig, ax = plt.subplots(figsize=(12,6))
ax.bar(x - w/2, counts1, w, label='No LC Filter', color='steelblue')
ax.bar(x + w/2, counts2, w, label='With LC Filter', color='green')
ax.set(xlabel='Chromosome', ylabel='Proportion with Close Seq in Chimp Gap Divergence Region',
title='Human Gap Divergence Example Seqs vs Matching Chimp Chrm Gap Divergence Seqs',
xticks=x, xticklabels=chroms)
plt.xticks(rotation=45, ha='right')
ax.legend()
plt.tight_layout()
plt.savefig('Fig3.png', dpi=600)
plt.show()
"""